光纤光度数据扩展
项目描述
ndx-photometry NWB扩展
介绍
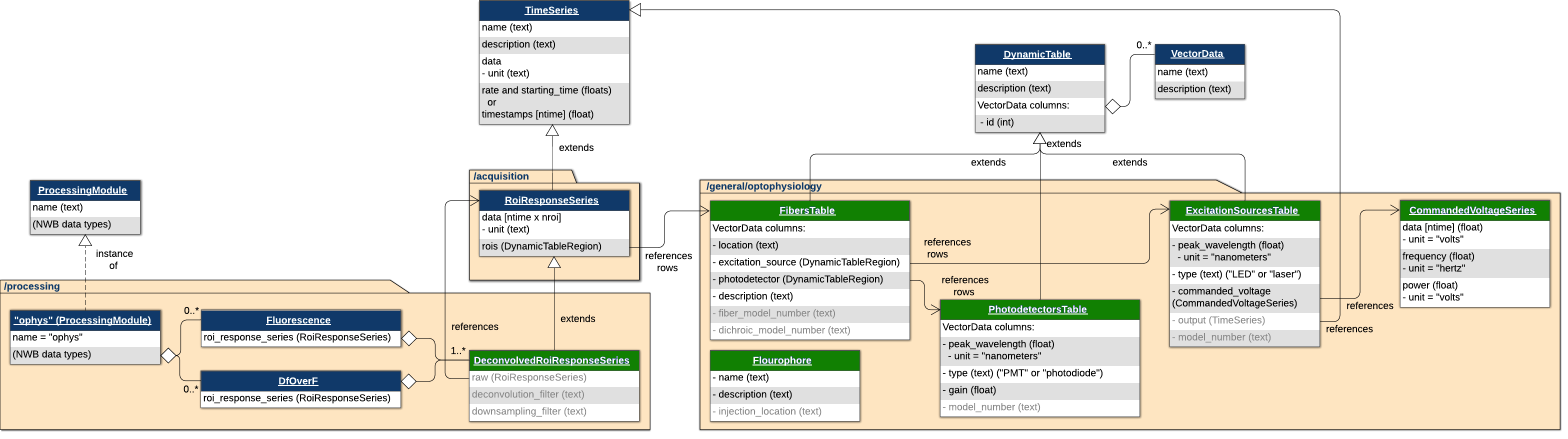
这是一个用于存储光度记录和相关元数据的NWB扩展。此扩展将在NWB文件中的三个文件夹中存储光度信息:获取、处理和通用。获取文件夹包含一个FiberPhotometryResponseSeries,它引用了FibersTable、ExcitationSourcesTable、PhotodetectorsTable和FluorophoresTable的行。此扩展的新类型在元数据和处理中。
元数据
FibersTable为每个光纤存储行,包含有关位置、光电探测器和更多信息(与每个光纤相关)。ExcitationSourcesTable为每个激发源存储行,包含有关峰值波长、源类型以及类型为CommandedVoltageSeries的命令电压序列。PhotodectorsTable为每个光电探测器存储行,包含有关峰值波长、类型等信息。FluorophoresTable为每个荧光素存储行,包含有关荧光素本身和注射部位的信息。
处理
DeconvoledROIResponseSeries存储DfOverF和荧光轨迹,并扩展了ROIResponseSeries以包含关于所执行的解卷积和下采样过程的信息。
此扩展由Akshay Jaggi、Ben Dichter和Ryan Ly开发。
安装
pip install ndx-photometry
用法
import datetime
import numpy as np
from pynwb import NWBHDF5IO, NWBFile
from pynwb.ophys import RoiResponseSeries
from ndx_photometry import (
FibersTable,
PhotodetectorsTable,
ExcitationSourcesTable,
FluorophoresTable,
FiberPhotometryResponseSeries,
FiberPhotometry
)
nwbfile = NWBFile(
session_description="session_description",
identifier="identifier",
session_start_time=datetime.datetime.now(datetime.timezone.utc),
)
# Create a Fibers table, and add one (or many) fiber
fibers_table = FibersTable(description="fibers table")
fibers_table.add_row(
location="my location",
notes="notes"
)
# Create an Excitation Sources table, and a one (or many) excitation source
excitationsources_table = ExcitationSourcesTable(description="excitation sources table")
excitationsources_table.add_row(
peak_wavelength=700.0,
source_type="laser",
)
# Create a Photodetectors table, and add one (or many) photodetector
photodetectors_table = PhotodetectorsTable(description="photodetectors table")
photodetectors_table.add_row(
peak_wavelength=500.0,
type="PMT",
gain=100.0
)
# Create a Fluorophores table, and add one (or many) fluorophore
fluorophores_table = FluorophoresTable(description="fluorophores")
fluorophores_table.add_row(
label="dlight",
location="VTA",
coordinates=(3.0,2.0,1.0),
excitation_peak_wavelength=700.0,
emission_peak_wavelength=500.0
)
# Here we add the metadata tables to the metadata section
nwbfile.add_lab_meta_data(
FiberPhotometry(
fibers=fibers_table,
excitation_sources=excitationsources_table,
photodetectors=photodetectors_table,
fluorophores=fluorophores_table
)
)
# Create a raw FiberPhotometryResponseSeries, this is your main acquisition
# We should create DynamicTableRegion referencing the correct rows for each table
fiber_ref = fibers_table.create_fiber_region(region=[0], description='source fiber')
excitation_ref = excitationsources_table.create_excitation_source_region(region=[0], description='excitation sources')
photodetector_ref = photodetectors_table.create_photodetector_region(region=[0], description='photodetector')
fluorophore_ref = fluorophores_table.create_fluorophore_region(region=[0], description='fluorophore')
fp_response_series = FiberPhotometryResponseSeries(
name="MyFPRecording",
data=np.random.randn(100, 1),
unit='F',
rate=30.0,
fibers=fiber_ref,
excitation_sources=excitation_ref,
photodetectors=photodetector_ref,
fluorophores=fluorophore_ref,
)
nwbfile.add_acquisition(fp_response_series)
# write nwb file
filename = 'test.nwb'
with NWBHDF5IO(filename, 'w') as io:
io.write(nwbfile)
# read nwb file and check its contents
with NWBHDF5IO(filename, 'r', load_namespaces=True) as io:
nwbfile = io.read()
# Access and print information about the acquisition
print(nwbfile.acquisition["MyFPRecording"])
# Access and print all of the metadata
print(nwbfile.lab_meta_data)
此扩展使用 ndx-template 创建。