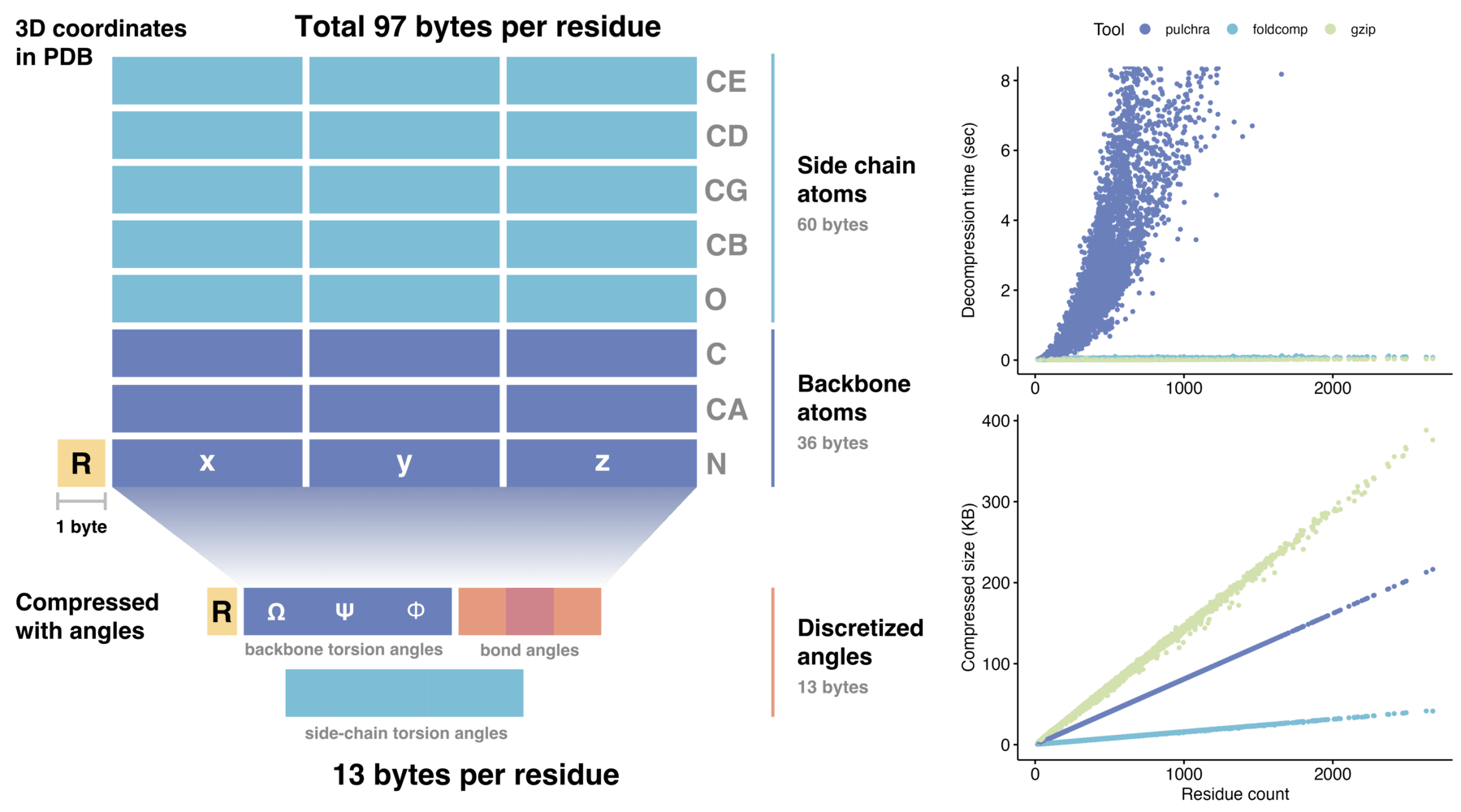
foldcomp通过有效地压缩蛋白质结构的扭转角。它将骨架原子压缩到8个字节,每个残基的侧链额外4-5个字节,一个平均大小的350个残基的蛋白质需要约4.2kb。Foldcomp是一个C++库,具有Python绑定。
项目描述
Foldcomp

Foldcomp高效的压缩格式存储蛋白质结构,每个残基只需13个字节,与直接保存3D坐标相比,所需存储空间减少了10倍。我们通过将骨架的扭转角以及侧链角度编码为紧凑的二进制文件格式(FCZ)来实现这种减少。
Foldcomp目前仅支持压缩单链PDB文件
出版物
使用方法
安装Foldcomp
# Install Foldcomp Python package
pip install foldcomp
# Download static binaries for Linux
wget https://mmseqs.com/foldcomp/foldcomp-linux-x86_64.tar.gz
# Download static binaries for Linux (ARM64)
wget https://mmseqs.com/foldcomp/foldcomp-linux-arm64.tar.gz
# Download binary for macOS
wget https://mmseqs.com/foldcomp/foldcomp-macos-universal.tar.gz
# Download binary for Windows (x64)
wget https://mmseqs.com/foldcomp/foldcomp-windows-x64.zip
可执行文件
# Compression
foldcomp compress <pdb|cif> [<fcz>]
foldcomp compress [-t number] <dir|tar(.gz)> [<dir|tar|db>]
# Decompression
foldcomp decompress <fcz|tar> [<pdb>]
foldcomp decompress [-t number] <dir|tar(.gz)|db> [<dir|tar>]
# Decompressing a subset of Foldcomp database
foldcomp decompress [-t number] --id-list <idlist.txt> <db> [<dir|tar>]
# Extraction of sequence or pLDDT
foldcomp extract [--plddt|--amino-acid] <fcz> [<fasta>]
foldcomp extract [--plddt|--amino-acid] [-t number] <dir|tar(.gz)|db> [<fasta_out>]
# Check
foldcomp check <fcz>
foldcomp check [-t number] <dir|tar(.gz)|db>
# RMSD
foldcomp rmsd <pdb|cif> <pdb|cif>
# Options
-h, --help print this help message
-v, --version print version
-t, --threads threads for (de)compression of folders/tar files [default=1]
-r, --recursive recursively look for files in directory [default=0]
-f, --file input is a list of files [default=0]
-a, --alt use alternative atom order [default=false]
-b, --break interval size to save absolute atom coordinates [default=25]
-z, --tar save as tar file [default=false]
-d, --db save as database [default=false]
-y, --overwrite overwrite existing files [default=false]
-l, --id-list a file of id list to be processed (only for database input)
--skip-discontinuous skip PDB with with discontinuous residues (only batch compression)
--check check FCZ before and skip entries with error (only for batch decompression)
--plddt extract pLDDT score (only for extraction mode)
--fasta extract amino acid sequence (only for extraction mode)
--no-merge do not merge output files (only for extraction mode)
--time measure time for compression/decompression
下载数据库
我们为多个大型预测蛋白质结构集合提供预构建的数据库,并有一个Python辅助程序来下载数据库文件。
您可以使用以下命令下载AlphaFoldDB Swiss-Prot
python -c "import foldcomp; foldcomp.setup('afdb_swissprot_v4');
目前我们提供的数据库有
-
ESMAtlas 完整版(v0 + v2023_02):
foldcomp.setup('esmatlas') -
ESMAtlas v2023_02:
foldcomp.setup('esmatlas_v2023_02') -
ESMAtlas 高质量:
foldcomp.setup('highquality_clust30')注意: 我们跳过了所有具有不连续残基或其他问题的结构。以下是受影响预测的列表;完整版 (~21M),高质量 (~100k),v2023_02 (~10k)
-
AlphaFoldDB Uniprot:
foldcomp.setup('afdb_uniprot_v4') -
AlphaFoldDB Swiss-Prot:
foldcomp.setup('afdb_swissprot_v4') -
AlphaFoldDB 模型生物:
foldcomp.setup('h_sapiens')a_thaliana,c_albicans,c_elegans,d_discoideum,d_melanogaster,d_rerio,e_coli,g_max,h_sapiens,m_jannaschii,m_musculus,o_sativa,r_norvegicus,s_cerevisiae,s_pombe,z_mays
-
AlphaFoldDB 集群代表:
foldcomp.setup('afdb_rep_v4') -
AlphaFoldDB 集群代表(暗群):
foldcomp.setup('afdb_rep_dark_v4')
如果您需要其他预构建的数据集,请通过我们的 GitHub 问题 与我们联系。
如果您在下载数据库时遇到问题,可以直接导航到我们的 下载服务器 并下载所需的文件。例如,afdb_uniprot_v4、afdb_uniprot_v4.index、afdb_uniprot_v4.dbtype、afdb_uniprot_v4.lookup 和可选的 afdb_uniprot_v4.source。
Python API
您可以在示例笔记本中找到更多关于使用 Foldcomp Python 接口的深入示例:[点击打开 Colab](https://colab.research.google.com/github/steineggerlab/foldcomp/blob/master/foldcomp-py-examples.ipynb)
import foldcomp
# 01. Handling a FCZ file
# Open a fcz file
with open("test/compressed.fcz", "rb") as fcz:
fcz_binary = fcz.read()
# Decompress
(name, pdb) = foldcomp.decompress(fcz_binary) # pdb_out[0]: file name, pdb_out[1]: pdb binary string
# Save to a pdb file
with open(name, "w") as pdb_file:
pdb_file.write(pdb)
# Get data as dictionary
data_dict = foldcomp.get_data(fcz_binary) # foldcomp.get_data(pdb) also works
# Keys: phi, psi, omega, torsion_angles, residues, bond_angles, coordinates
data_dict["phi"] # phi angles (C-N-CA-C)
data_dict["psi"] # psi angles (N-CA-C-N)
data_dict["omega"] # omega angles (CA-C-N-CA)
data_dict["torsion_angles"] # torsion angles of the backbone as list (phi + psi + omega)
data_dict["bond_angles"] # bond angles of the backbone as list
data_dict["residues"] # amino acid residues as string
data_dict["coordinates"] # coordinates of the backbone as list
# 02. Iterate over a database of FCZ files
# Open a foldcomp database
ids = ["d1asha_", "d1it2a_"]
with foldcomp.open("test/example_db", ids=ids) as db:
# Iterate through database
for (name, pdb) in db:
# save entries as seperate pdb files
with open(name + ".pdb", "w") as pdb_file:
pdb_file.write(pdb)
数据库子集
如果您正在处理数百万条条目,我们建议使用 mmseqs2 的 createsubdb 命令对数据库进行子集处理。以下命令可以用于根据给定 ID 子集 AlphaFold Uniprot DB。
# mmseqs createsubdb --subdb-mode 0 --id-mode 1 id_list.txt input_foldcomp_db output_foldcomp_db
mmseqs createsubdb --subdb-mode 0 --id-mode 1 id_list.txt afdb_uniprot_v4 afdb_subset
请注意,afdb_uniprot_v4 中的 ID 格式为 AF-A0A5S3Y9Q7-F1-model_v4。
社区贡献
- PyMOL 插件用于读取 Foldcomp 文件 by @yakomaxa
贡献者

项目详情
下载文件
下载适合您平台的文件。如果您不确定选择哪个,请了解更多关于 安装包 的信息。
源代码发行版
构建发行版
哈希值 for foldcomp-0.0.7-cp311-cp311-musllinux_1_1_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 1946c813087de71841c92f2763b0926ce9e89f5f1882a3ed49fa6b85880921c1 |
|
| MD5 | 851e935ba702098d1e5cb7941b29458a |
|
| BLAKE2b-256 | 702f80814f26928bbffd89bc1d2ea9143b52797520fd7b831b0de12940366d82 |
哈希值 for foldcomp-0.0.7-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 52cac8d0e9c48369ffb1f7848961afd900fe0fddca3cb07427809969cf39f575 |
|
| MD5 | e4aa3847f4cfeec42855da4faa8cb422 |
|
| BLAKE2b-256 | ee40dc68eba8e424740c1610928631653acc3f58b01f9cca26bd6e81ab7073ac |
哈希值 for foldcomp-0.0.7-cp311-cp311-macosx_10_9_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | b279f5ad06193b78db7fe680b05f53809e6e97f630356d3dc57ac1f5528ae8bc |
|
| MD5 | ec4b09032a829cc06cf6b711fee9e397 |
|
| BLAKE2b-256 | 6e896ccaeb1ebd392c4bd622d1c646ccc2ecf324b6e88f04dd7278d230055ab2 |
哈希值 for foldcomp-0.0.7-cp311-cp311-macosx_10_9_universal2.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 7aec763b930f2552871b925e043173417dfd7920accaa4e823a2eb1eb30054fe |
|
| MD5 | ddcb7225912f3ed7517a3837cb02b8b0 |
|
| BLAKE2b-256 | 2a00688fc1bfac5b1996b36a4f3e851a4198fbae8266981431e9161c16afd382 |
哈希值 for foldcomp-0.0.7-cp310-cp310-musllinux_1_1_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 218720c0ce3f0fa7088ca30bfff4ec34ef83f3afbea9b87fb9426bbee1e6c30c |
|
| MD5 | 21a14e21ea4b14c6b3022f83699de3aa |
|
| BLAKE2b-256 | b25ad95a18c8ec525ab7727d0557042c719e13e2d791bce6f3cfb5f284936ac9 |
哈希值 for foldcomp-0.0.7-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | ff229be20e2cfffa12b5c566a4c92c9667b1546300b5669c25c21baf58cd0e4e |
|
| MD5 | 16092ecbf281190604bbfe7b956ac6c1 |
|
| BLAKE2b-256 | 33fd9f7bb10b8925750e728456af8a869ae713273e74a04afa47f749b4e5c59d |
哈希值 for foldcomp-0.0.7-cp310-cp310-macosx_10_9_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 8174d9d4aa43ab1e3f012b6b6eaeb1d4bab23ef7a8b9e931ae69e0c4579cb5c1 |
|
| MD5 | b34e844f0130d76e1f41c26ff62b8e2f |
|
| BLAKE2b-256 | a4dd4204d433ebf069e542c05fa870c8339011dcae2491875ebfa04b736f8bd9 |
哈希值 for foldcomp-0.0.7-cp310-cp310-macosx_10_9_universal2.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 780cf8abf687d25822c3d393b790540adef8f246cb226804962a79a351b95b57 |
|
| MD5 | df28d4bfb6431a9adcf401d893c9f4db |
|
| BLAKE2b-256 | e7468b306556de097b31e2cf11ead0f3f15bcf6a84fda3e8004e3d8a08df7a0c |
哈希值 for foldcomp-0.0.7-cp39-cp39-musllinux_1_1_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 009956e7678045a7f49e2fa7c5ea30311090107ae0f562dd8bd4037fd53fdb32 |
|
| MD5 | 5fc7786a896f7f3ae2066c1cf4c3e1de |
|
| BLAKE2b-256 | 679d4d6d3f03525f173380abb347e12b7c62b7006583c614079742e4f8dd90c9 |
哈希值 用于 foldcomp-0.0.7-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | f41aff4c18399811a2571f3d999a9abb33c681fcb0cbb96d87f3c2f7616235bc |
|
| MD5 | 071341c4502efe8ea737da3c62aca863 |
|
| BLAKE2b-256 | ece69f70dfae43c2afc1b197dcae2d80006550f29a44563a8776c5ae704321f1 |
哈希值 用于 foldcomp-0.0.7-cp39-cp39-macosx_10_9_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 83401f32dc996ec26314691ab5e65ed362d4adcd2c2935cbef2996cdf47e8038 |
|
| MD5 | 144c95c616c67bca4869bdf4097b46c7 |
|
| BLAKE2b-256 | f55bf783e68dce29530a192e23b8111f373a3128fe9532b63df9a4d2653036a0 |
哈希值 用于 foldcomp-0.0.7-cp39-cp39-macosx_10_9_universal2.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 24eb2ba08e6fe908e4b239a6ba590ec54d1b851620e419744df6e697bdf36a66 |
|
| MD5 | e7e97b2e5fd46bd4fe30baf2e02b0e97 |
|
| BLAKE2b-256 | 04ca0aa8d6f325610ec0054b9e89d8deab7560f33a3e1345a208deb48b4685d9 |
哈希值 用于 foldcomp-0.0.7-cp38-cp38-musllinux_1_1_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 0c797e533570f1e03ce926200de50652880c4d1b266c12d8b297a2e7f16827c8 |
|
| MD5 | 36f1b228d21bc704b5efee8f6cbeea08 |
|
| BLAKE2b-256 | 5a65f373340cb0565e660501b59bc8007e3c14b86966d424d4e07ef9b5dcf23d |
哈希值 用于 foldcomp-0.0.7-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 8e3efee2e1ba4661ed376b6436ba88bd0c90037698de5d4085fe6ab64dfb552f |
|
| MD5 | 366d02fa007e92dc2dbc5c090bdc172c |
|
| BLAKE2b-256 | 00535e0f03e50e2dc0c07f99daf21d9edd68926a16357d4e9be0b3d1ceedc203 |
哈希值 用于 foldcomp-0.0.7-cp38-cp38-macosx_10_9_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | facaf4d301ab49dc97658dd80939995f8722f9c21a66ddc39024fb692ea8d5e6 |
|
| MD5 | 0d6384c3e585d8c52c92c851dfa9cfa9 |
|
| BLAKE2b-256 | 3d1dc1b0a634f6c1c9469acad47983cc2ad282dbd8d852b979e12a345e57cf70 |
哈希值 用于 foldcomp-0.0.7-cp38-cp38-macosx_10_9_universal2.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | bfbaf1f60fd523067b51bdada473060d9146848c0efb78d39109ac9fed572707 |
|
| MD5 | f2284b733af0fe6d4d379080751d4439 |
|
| BLAKE2b-256 | d7be5c2a9fb830afa3be5910b0933c80c6e1f97f10018207da027d5673f92c70 |
哈希值 用于 foldcomp-0.0.7-cp37-cp37m-musllinux_1_1_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | be7daf7e0c203f81953c209372655427e1800f140746ea569d0b861ae8883f9d |
|
| MD5 | 05a6d560de0b75147db7fcfa372a363a |
|
| BLAKE2b-256 | 934188753c31bef3d42666ed4efb45fe0f1308a345358d5966165f1c8347ef89 |
哈希值 用于 foldcomp-0.0.7-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 6e57601144439646f9a28fc1a981b8dfd4498d7e7630e24dd9eee0cd8295a3f9 |
|
| MD5 | 6ad9fec25ba5d39708cdd8448e11cefb |
|
| BLAKE2b-256 | 005dcd94c0bb832720fa1357cd155e267d60c4719e3c6c0f2ae2b3a98b1ddc58 |
哈希值 用于 foldcomp-0.0.7-cp37-cp37m-macosx_10_9_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | 629270d531c2187307ba6159af6085808ea646565b67d03718bda5109953b9b1 |
|
| MD5 | 88d66a3f629b5ef880941f0eb628be8b |
|
| BLAKE2b-256 | 285380f9a776b06439e28296a2111e43362d4a4511d76811bde192da0b2ab0f2 |











