Dynamask - 使用动态掩码解释时间序列预测
项目描述
Dynamask - 使用动态掩码解释时间序列预测
代码作者:Jonathan Crabbé (jc2133@cam.ac.uk)
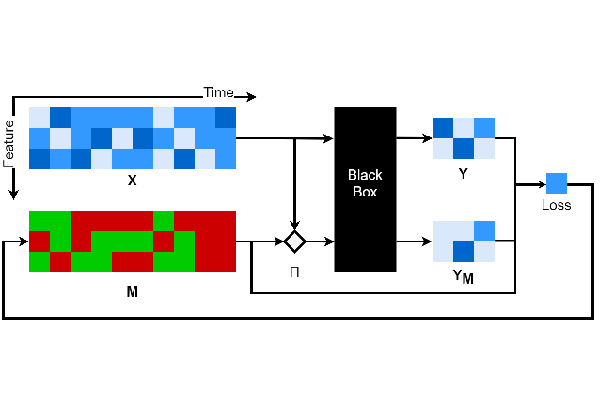
本存储库包含Dynamask的实现,该方法用于识别在数据以时间序列表示时,模型发出预测的显著特征。有关理论方面的更多详情,请阅读我们的ICML 2021论文:“使用动态掩码解释时间序列预测”。
我们论文中的部分实验依赖于FIT,这是另一个与NeurIPS 2021论文:“What went wrong and when? Instance-wise feature importance for time-series black-box models”相关的存储库。我们已在fit文件夹中包含了所有相关文件。
:rocket: 安装
该库需要libpq-dev。
您可以使用以下方法从PyPI安装库
$ pip install dynamask
或者从源安装,使用
$ pip install .
示例
在时间序列模型上拟合掩码非常容易。以下是一个拟合掩码到输入时间序列的玩具演示。在这种情况下,掩码区域固定为0.1(通过掩码突出显示的最重要的10%特征)。所有相关代码均可在文件mask中找到。
import torch
from dynamask.attribution.mask import Mask
from dynamask.attribution.perturbation import GaussianBlur
from dynamask.utils.losses import mse
torch.manual_seed(42)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
# Define a pseudo-black box:
def black_box(input):
output = input[-1, :] # The black-box returns the features of the last time step
return output
# Define a random input:
X = torch.randn(10, 3).to(device) # The shape of the input has to be (T, N_features)
# Fit a mask to the input with a Gaussian Blur perturbation:
pert = GaussianBlur(device)
mask = Mask(pert, device)
mask.fit(X, black_box, loss_function=mse, keep_ratio=0.1, size_reg_factor_init=0.01) # Select the 10% most important features
# Plot the resulting saliency map:
mask.plot_mask()
如果不知道要选择的特征的比率,一个好的方法是拟合一组具有不同面积的面具。然后,可以从这组中提取出极值面具。相关代码可以在文件mask_group中找到。
import torch
from dynamask.attribution.mask_group import MaskGroup
from dynamask.attribution.perturbation import GaussianBlur
from dynamask.utils.losses import mse
torch.manual_seed(42)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
# Define a pseudo-black box:
def black_box(input):
output = input[-1, :] # The black-box returns the features of the last time step
return output
# Define a random input:
X = torch.randn(10, 3).to(device) # The shape of the input has to be (T, N_features)
# Fit a group of masks to the input with a Gaussian Blur perturbation:
areas = [.1, .15, .2, .25] # These are the areas of the different masks
pert = GaussianBlur(device)
masks = MaskGroup(pert, device)
masks.fit(X, black_box, loss_function=mse, area_list=areas, size_reg_factor_init=0.01)
# Extract the extremal mask:
epsilon = 0.01
mask = masks.get_extremal_mask(threshold=epsilon)
# Plot the resulting saliency map:
mask.plot_mask()
复制实验
ICML论文中的所有实验都可以轻松复制。必要的代码在experiments中。下面,我们详细说明每个实验的步骤。
还提供了运行实验的脚本:experiments/run_<EXPERIMENT>.sh。
复制Rare实验
- 从存储库文件夹运行以下命令
python -m dynamask.experiments.rare_feature # Runs the Rare Feature experiment python -m dynamask.experiments.rare_time # Runs the Rare Time experiment
要使用各种种子进行实验,请将这些命令添加以下指定Options: --cv # An integer that sets the random seed (first run cv=0, second run cv=1, ...)
- 这些实验的结果保存在以下两个文件夹中:Rare Feature和Rare Time。要处理结果并计算相关指标,请运行
python -m dynamask.experiments.results.rare_feature.get_results python -m dynamask.experiments.results.rare_time.get_results
以下选项需要指定Options: --CV # The number of runs you have done for the experiment --explainers # The baselines you have used among: dynamask, fo, fp, ig, shap (separated by a space)
复制State实验
-
运行此命令以生成合成数据并将其存储在
data/state中python -m dynamask.fit.data_generator.state_data --signal_len 200 --signal_num 1000
-
运行以下命令以拟合一个模型和一个基线显著性方法
python -m dynamask.fit.evaluation.baselines --explainer fit --train
要使用各种基线进行实验,请更改解释器
Options: --explainer # The baselines can be: fit, lime, retain, integrated_gradient, deep_lift, fo, afo, gradient_shap --train # Only put this option when fitting the FIRST baseline (this is to avoid retraining a model for each baseline); however, required for retain baseline --cv # An integer that sets the random seed (first run cv=0, second run cv=1, ...)
-
模型和基线显著性图都保存在此文件夹中。现在,通过运行以下命令为每个时间序列拟合一个面具
python -m dynamask.experiments.state
请使用与上一个命令相同的
--cv选项。 -
面具都保存在此文件夹中。要处理结果并计算相关指标,请运行
python -m dynamask.experiments.results.state.get_results
以下选项需要指定
Options: --CV # The number of runs you have done for the experiment --explainers # The baselines you have used among: dynamask, fo, afo, deep_lift, fit, gradient_shap, integrated_gradient, lime, retain (separated by a space)
复制MIMIC实验
-
MIMIC-III是一个私有数据集。对于以下内容,您需要在本地服务器上运行MIMIC-III数据库。有关更多信息,请参阅官方MIMIC-III文档。
-
运行此命令以获取数据并将其存储
python dynamask/fit/data_generator/icu_mortality.py --sqluser YOUR_USER --sqlpass YOUR_PASSWORD
如果一切正常,两个名为
adult_icu_vital.gz和adult_icu_lab.gz的文件将存储在data/mimic中。 -
运行此命令以预处理数据
python dynamask/fit/data_generator/data_preprocess.py如果一切正常,一个名为
patient_vital_preprocessed.pkl的文件将存储在data/mimic中。 -
运行以下命令以拟合一个模型和一个基线显著性方法
python -m dynamask.fit.evaluation.baselines --data mimic --explainer fit --train
要使用各种基线进行实验,请更改解释器
Options: --explainer # The baselines can be: fit, lime, retain, integrated_gradient, deep_lift, fo, afo, gradient_shap --train # Only put this option when fitting the FIRST baseline (this is to avoid retraining a model for each baseline); however, required for retain baseline --cv # An integer that sets the random seed (first run cv=0, second run cv=1, ...)
-
模型和基线显著性图都保存在此文件夹中。现在,通过运行以下命令为每个时间序列拟合一个面具
python -m dynamask.experiments.mimic
请使用与上一个命令相同的
--cv选项。Options: --cv # Same as in the previous command --area # The area of the mask to fit (a number between 0 and 1)
-
面具都保存在此文件夹中。要处理结果并计算相关指标,请运行
python -m dynamask.experiments.results.state.plot_benchmarks
以下选项需要指定
Options: --CV # The number of runs you have done for the experiment --explainers # The baselines you have used among: dynamask, fo, afo, deep_lift, fit, gradient_shap, integrated_gradient, lime, retain (separated by a space) --areas # The mask areas that you have computed (separated by a space)
生成的图保存在此文件夹中。
:hammer: 测试
使用以下命令安装测试依赖项
pip install .[testing]
可以使用以下命令执行测试
pytest -vsx
引用
如果您使用此代码,请引用相关论文
@InProceedings{pmlr-v139-crabbe21a,
title = {Explaining Time Series Predictions with Dynamic Masks},
author = {Crabb{\'e}, Jonathan and Van Der Schaar, Mihaela},
booktitle = {Proceedings of the 38th International Conference on Machine Learning},
pages = {2166--2177},
year = {2021},
editor = {Meila, Marina and Zhang, Tong},
volume = {139},
series = {Proceedings of Machine Learning Research},
month = {18--24 Jul},
publisher = {PMLR},
pdf = {http://proceedings.mlr.press/v139/crabbe21a/crabbe21a.pdf},
url = {https://proceedings.mlr.press/v139/crabbe21a.html},
abstract = {How can we explain the predictions of a machine learning model? When the data is structured as a multivariate time series, this question induces additional difficulties such as the necessity for the explanation to embody the time dependency and the large number of inputs. To address these challenges, we propose dynamic masks (Dynamask). This method produces instance-wise importance scores for each feature at each time step by fitting a perturbation mask to the input sequence. In order to incorporate the time dependency of the data, Dynamask studies the effects of dynamic perturbation operators. In order to tackle the large number of inputs, we propose a scheme to make the feature selection parsimonious (to select no more feature than necessary) and legible (a notion that we detail by making a parallel with information theory). With synthetic and real-world data, we demonstrate that the dynamic underpinning of Dynamask, together with its parsimony, offer a neat improvement in the identification of feature importance over time. The modularity of Dynamask makes it ideal as a plug-in to increase the transparency of a wide range of machine learning models in areas such as medicine and finance, where time series are abundant.}
}
项目详情
下载文件
下载适合您平台的文件。如果您不确定选择哪个,请了解有关安装包的更多信息。
源代码分发
构建分发
哈希值 for dynamask-0.0.1-py3-none-macosx_10_14_x86_64.whl
| 算法 | 哈希摘要 | |
|---|---|---|
| SHA256 | ea7f93ac573b12a4732d3add3db39b50000e31dbf1daf4df3be106bf170e6bc0 |
|
| MD5 | 60c50b9ae62ab870816ae35dd32f441f |
|
| BLAKE2b-256 | 9360d7091ee661edbc0519ed6ae86161f3b0e3d1f9a3458148b31ccfa3292008 |